記述統計#
データの要約#
次のようなデータが与えられたとする.
データを眺めていても傾向は読めないので,np.sort関数を使ってデータを順に並べてみる
import numpy as np
X = np.array([4, 1, 7, 6, 5, 6, 5, 4, 4, 3, 4, 4, 5, 5, 5, 5, 6, 6, 6, 5, 4, 5, 10])
Y = np.array([9, 6, 1, 6, 5, 10, 3, 2, 7, 2, 5, 8, 7, 6, 1, 8, 4, 5, 4, 0, 9, 3, 4])
print("X :=", np.sort(X) )
print("Y :=", np.sort(Y) )
X := [ 1 3 4 4 4 4 4 4 5 5 5 5 5 5 5 5 6 6 6 6 6 7 10]
Y := [ 0 1 1 2 2 3 3 4 4 4 5 5 5 6 6 6 7 7 8 8 9 9 10]
すると最小値や最大値が一目でわかり
print( np.min(X), np.max(X) ) # = X.max(), X.min()
print( np.min(Y), np.max(Y) ) # = Y.max(), Y.min()
1 10
0 10
もう少し詳しい情報をpandasパッケージを用いて要約させると
import pandas as pd
df = pd.DataFrame({
'試験X': X,
'試験Y': Y,
})
df.describe()
| 試験X | 試験Y | |
|---|---|---|
| count | 23.000000 | 23.000000 |
| mean | 5.000000 | 5.000000 |
| std | 1.651446 | 2.796101 |
| min | 1.000000 | 0.000000 |
| 25% | 4.000000 | 3.000000 |
| 50% | 5.000000 | 5.000000 |
| 75% | 6.000000 | 7.000000 |
| max | 10.000000 | 10.000000 |
とわかる.mean以下の詳細は次節で詳細に説明するが,ここではdf.discribe()の出力結果を簡潔に説明する.
まず,countはデータセットの大きさ(要素数,総度数)であり試験X,試験Yともに23個である. meanはデータセットの平均値であり試験X,試験Yともに5点である. stdは標準偏差であり,試験Xは約1.65点,試験Yは約2.80点である. 標準偏差はデータの散らばりを表す指標(散布度)であり,値が大きいほどデータのばらつきが大きいことを意味し,試験Yの方がデータが散らばっていうることがわかる(逆に試験Xは平均値の周りにデータが集まっている). minはデータセットの最小値, 25%,50%,75% はデータセットの25%点,50%点(中央値),75%点, maxはデータセットの最大値を意味する. このようにデータセットの特徴量を要約することで,データの傾向を把握することができる.
代表値#
n = len(X)
print("平均値 :", np.sum(X) / n )
print("平均値 :", np.mean(X) )
平均値 : 5.0
平均値 : 5.0
X = np.array([50, 100, 150, 200, 4500])
print("平均値:", np.mean(X) )
print("中央値:", np.median(X) )
平均値: 1000.0
中央値: 150.0
散布度#
平均偏差#
X = np.array([4, 1, 7, 6, 5, 6, 5, 4, 4, 3, 4, 4, 5, 5, 5, 5, 6, 6, 6, 5, 4, 5, 10])
Dx = X - X.mean()
print("Dx = ", Dx)
print("np.mean(Dx) = ", np.mean(Dx) )
Dx = [-1. -4. 2. 1. 0. 1. 0. -1. -1. -2. -1. -1. 0. 0. 0. 0. 1. 1.
1. 0. -1. 0. 5.]
np.mean(Dx) = 0.0
問 Yのデータセットに対して同様に偏差の平均値が0になることを示せ.
Y = np.array([9, 6, 1, 6, 5, 10, 3, 2, 7, 2, 5, 8, 7, 6, 1, 8, 4, 5, 4, 0, 9, 3, 4])
2乗偏差と分散・標準偏差#
X = np.array([4, 1, 7, 6, 5, 6, 5, 4, 4, 3, 4, 4, 5, 5, 5, 5, 6, 6, 6, 5, 4, 5, 10])
Dx = X - X.mean() # = [𝑑_1, ... , 𝑑_𝑛] where 𝑑𝑖 = (𝑥_𝑖 - 𝑥)
Dx2 = Dx ** 2 # = [𝑑^2_1 , ... ,𝑑^2_𝑛] where 𝑑^2_𝑖 = (𝑥_𝑖 - 𝑥)^2
print("Dx2 = ", Dx2)
print("np.mean(Dx2) = ", np.mean(Dx2) )
Dx2 = [ 1. 16. 4. 1. 0. 1. 0. 1. 1. 4. 1. 1. 0. 0. 0. 0. 1. 1.
1. 0. 1. 0. 25.]
np.mean(Dx2) = 2.608695652173913
print( X.var() )
2.608695652173913
問 Yのデータセットに対して同様に2乗偏差の平均値を求め,np.var(Y)の値と一致することを示せ.
print("std = ", np.sqrt( np.var(X) ))
std = 1.6151457061744965
print(r"std = ", np.std(X) ) # = X.std()
std = 1.6151457061744965
X2 = X**2
分散の計算公式#
\(s^2 = \overline{(x^2)} - \overline{(x)}^2\)から分散の値を求める.
X = np.array([4, 1, 7, 6, 5, 6, 5, 4, 4, 3, 4, 4, 5, 5, 5, 5, 6, 6, 6, 5, 4, 5, 10])
X2 = X ** 2 # $= [4^2, 1^2, ..., 5^2, 10^2]$
s2 = np.mean(X2) - np.mean(X) ** 2 # $= ¥overline{(x^2)} - (¥overline{x})^2$
print( s2 )
print( X.var() )
2.608695652173914
2.608695652173913
問 データセット\(Y\)に対して同様に,\(Y^2\)のデータセットを作成し,分散の計算公式から分散\(s^2\)を評価せよ.またその値がnp.var(Y)の値に一致することを確認せよ.
データの整理#
名義尺度の場合#
import pandas as pd
import numpy as np
X = ["A", "B", "O", "A", "O", "AB", "B", "O", "A", "O",
"A", np.nan, "B", "AB", "O", "O", "A", "O", "B", "AB", "O"]
bloodtypes = pd.Series(X)
print("unique dataset =", bloodtypes.unique() )
print("総度数 =", bloodtypes.count() )
print("Xの大きさ = ", len(X) )
unique dataset = ['A' 'B' 'O' 'AB' nan]
総度数 = 20
Xの大きさ = 21
bloodtypes = pd.Series(X).dropna()
print("unique dataset =", bloodtypes.unique() )
unique dataset = ['A' 'B' 'O' 'AB']
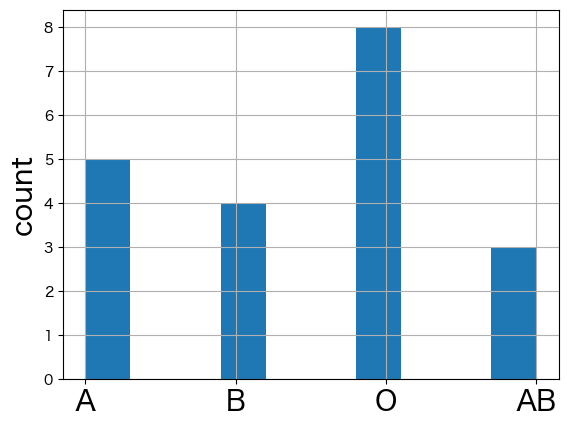
bloodtypes.value_counts().reindex(["A", "B", "O", "AB"])
A 5
B 4
O 8
AB 3
Name: count, dtype: int64
import matplotlib.pyplot as plt
plt.ylabel("count", fontsize=20)
bloodtypes.hist(xlabelsize=20)
<Axes: ylabel='count'>
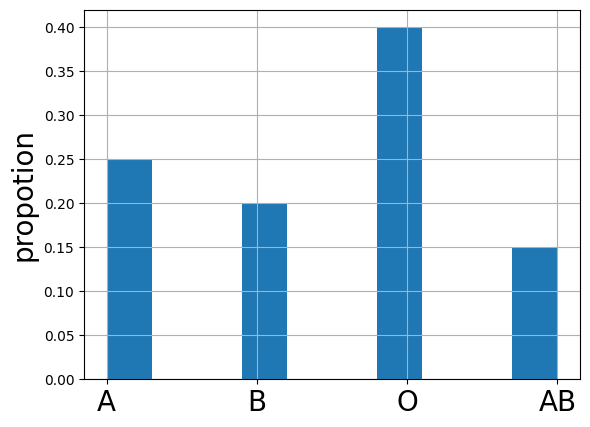
bloodtypes.value_counts(normalize=True).reindex(["A", "B", "O", "AB"])
| proportion | |
|---|---|
| A | 0.25 |
| B | 0.20 |
| O | 0.40 |
| AB | 0.15 |
n = bloodtypes.count()
w = np.ones(n) / n
print(w)
[0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05
0.05 0.05 0.05 0.05 0.05 0.05]
plt.ylabel("propotion",fontsize=20)
bloodtypes.hist(weights=w, xlabelsize=20)
<Axes: ylabel='propotion'>
量的データかつ離散データの場合#
data = np.array([4, 1, 7, 6, 5, 6, 5, 4, 4, 3, 4, 4, 5, 5, 5, 5, 6, 6, 6, 5, 4, 5, 10])
X = pd.Series(data)
X.value_counts()
| count | |
|---|---|
| 5 | 8 |
| 4 | 6 |
| 6 | 5 |
| 7 | 1 |
| 1 | 1 |
| 3 | 1 |
| 10 | 1 |
Xe = np.arange(0, 11) # = [0, 1, ..., 10]
X.value_counts().reindex(Xe)
| count | |
|---|---|
| 0 | NaN |
| 1 | 1.0 |
| 2 | NaN |
| 3 | 1.0 |
| 4 | 6.0 |
| 5 | 8.0 |
| 6 | 5.0 |
| 7 | 1.0 |
| 8 | NaN |
| 9 | NaN |
| 10 | 1.0 |
X.value_counts().reindex(Xe, fill_value=0)
| count | |
|---|---|
| 0 | 0 |
| 1 | 1 |
| 2 | 0 |
| 3 | 1 |
| 4 | 6 |
| 5 | 8 |
| 6 | 5 |
| 7 | 1 |
| 8 | 0 |
| 9 | 0 |
| 10 | 1 |
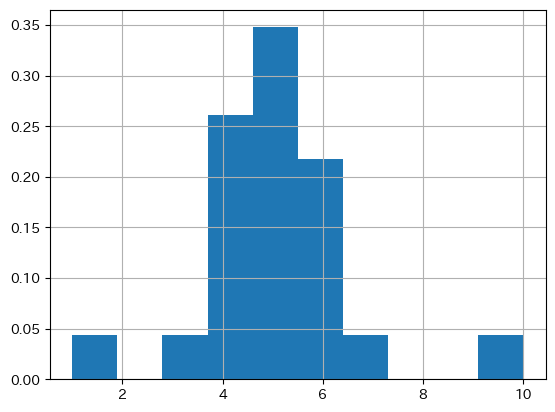
X.value_counts(normalize=True).reindex(Xe, fill_value=0)
| proportion | |
|---|---|
| 0 | 0.000000 |
| 1 | 0.043478 |
| 2 | 0.000000 |
| 3 | 0.043478 |
| 4 | 0.260870 |
| 5 | 0.347826 |
| 6 | 0.217391 |
| 7 | 0.043478 |
| 8 | 0.000000 |
| 9 | 0.000000 |
| 10 | 0.043478 |
Xe = np.arange(0, 11) # 0, 1, ..., 10
X.value_counts(normalize=True).reindex(Xe, fill_value=0).cumsum()
| proportion | |
|---|---|
| 0 | 0.000000 |
| 1 | 0.043478 |
| 2 | 0.043478 |
| 3 | 0.086957 |
| 4 | 0.347826 |
| 5 | 0.695652 |
| 6 | 0.913043 |
| 7 | 0.956522 |
| 8 | 0.956522 |
| 9 | 0.956522 |
| 10 | 1.000000 |
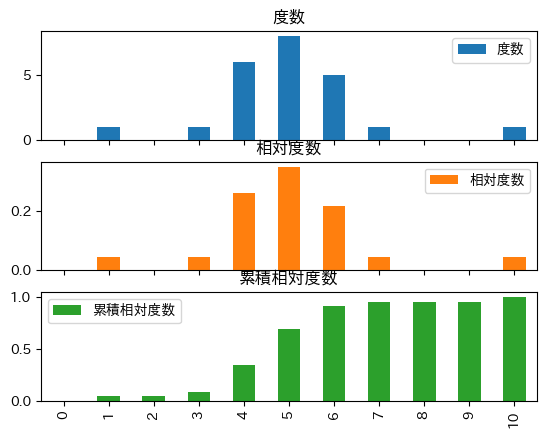
import numpy as np
import pandas as pd
data = np.array([4, 1, 7, 6, 5, 6, 5, 4, 4, 3, 4, 4, 5, 5, 5, 5, 6, 6, 6, 5, 4, 5, 10])
X = pd.Series(data)
Xe = np.arange(0, 11) # = [0, 1, ..., 10]
freq_tab = pd.DataFrame({
'度数': X.value_counts().reindex(Xe, fill_value=0),
'相対度数': X.value_counts(normalize=True).reindex(Xe, fill_value=0),
'累積相対度数': X.value_counts(normalize=True).reindex(Xe, fill_value=0).cumsum()
}, index = Xe)
freq_tab
| 度数 | 相対度数 | 累積相対度数 | |
|---|---|---|---|
| 0 | 0 | 0.000000 | 0.000000 |
| 1 | 1 | 0.043478 | 0.043478 |
| 2 | 0 | 0.000000 | 0.043478 |
| 3 | 1 | 0.043478 | 0.086957 |
| 4 | 6 | 0.260870 | 0.347826 |
| 5 | 8 | 0.347826 | 0.695652 |
| 6 | 5 | 0.217391 | 0.913043 |
| 7 | 1 | 0.043478 | 0.956522 |
| 8 | 0 | 0.000000 | 0.956522 |
| 9 | 0 | 0.000000 | 0.956522 |
| 10 | 1 | 0.043478 | 1.000000 |
!pip install japanize-matplotlib
import japanize_matplotlib
Requirement already satisfied: japanize-matplotlib in /usr/local/lib/python3.11/dist-packages (1.1.3)
Requirement already satisfied: matplotlib in /usr/local/lib/python3.11/dist-packages (from japanize-matplotlib) (3.10.0)
Requirement already satisfied: contourpy>=1.0.1 in /usr/local/lib/python3.11/dist-packages (from matplotlib->japanize-matplotlib) (1.3.2)
Requirement already satisfied: cycler>=0.10 in /usr/local/lib/python3.11/dist-packages (from matplotlib->japanize-matplotlib) (0.12.1)
Requirement already satisfied: fonttools>=4.22.0 in /usr/local/lib/python3.11/dist-packages (from matplotlib->japanize-matplotlib) (4.58.4)
Requirement already satisfied: kiwisolver>=1.3.1 in /usr/local/lib/python3.11/dist-packages (from matplotlib->japanize-matplotlib) (1.4.8)
Requirement already satisfied: numpy>=1.23 in /usr/local/lib/python3.11/dist-packages (from matplotlib->japanize-matplotlib) (2.0.2)
Requirement already satisfied: packaging>=20.0 in /usr/local/lib/python3.11/dist-packages (from matplotlib->japanize-matplotlib) (24.2)
Requirement already satisfied: pillow>=8 in /usr/local/lib/python3.11/dist-packages (from matplotlib->japanize-matplotlib) (11.2.1)
Requirement already satisfied: pyparsing>=2.3.1 in /usr/local/lib/python3.11/dist-packages (from matplotlib->japanize-matplotlib) (3.2.3)
Requirement already satisfied: python-dateutil>=2.7 in /usr/local/lib/python3.11/dist-packages (from matplotlib->japanize-matplotlib) (2.9.0.post0)
Requirement already satisfied: six>=1.5 in /usr/local/lib/python3.11/dist-packages (from python-dateutil>=2.7->matplotlib->japanize-matplotlib) (1.17.0)
freq_tab.plot(kind='bar', subplots=True)
array([<Axes: title={'center': '度数'}>, <Axes: title={'center': '相対度数'}>,
<Axes: title={'center': '累積相対度数'}>], dtype=object)
X.hist() # 度数
<Axes: >
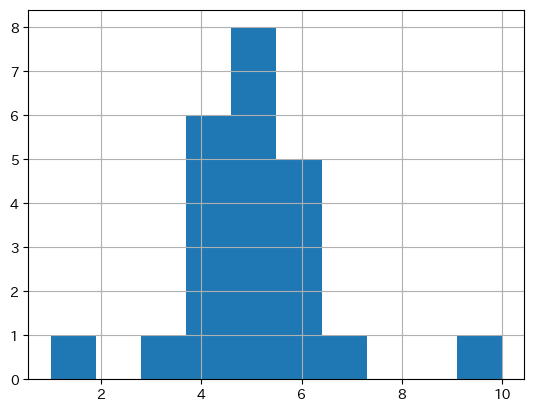
w = np.ones(X.count()) / X.count()
X.hist(weights=w) # 相対度数
<Axes: >
量的データかつ連続データの場合#
data = [161.05, 176.75, 167.71, 180.49, 176.56, 165.79, 163.04,
174.24, 171.99, 172.79, 163.93, 170.52, 173.68, 171.79, 164.07]
X = pd.Series(data)
X.value_counts()
| count | |
|---|---|
| 161.05 | 1 |
| 176.75 | 1 |
| 167.71 | 1 |
| 180.49 | 1 |
| 176.56 | 1 |
| 165.79 | 1 |
| 163.04 | 1 |
| 174.24 | 1 |
| 171.99 | 1 |
| 172.79 | 1 |
| 163.93 | 1 |
| 170.52 | 1 |
| 173.68 | 1 |
| 171.79 | 1 |
| 164.07 | 1 |
k = 5 # 階級の数
X.value_counts(bins=k).sort_index() # sort_index()をつけない場合は度数の大きい順に並ぶ
| count | |
|---|---|
| (161.03, 164.938] | 4 |
| (164.938, 168.826] | 2 |
| (168.826, 172.714] | 3 |
| (172.714, 176.602] | 4 |
| (176.602, 180.49] | 2 |
X.value_counts(bins=[160, 165, 170, 175, 180, 185]).sort_index()
| count | |
|---|---|
| (159.999, 165.0] | 4 |
| (165.0, 170.0] | 2 |
| (170.0, 175.0] | 6 |
| (175.0, 180.0] | 2 |
| (180.0, 185.0] | 1 |
import pandas as pd
data = [161.05, 176.75, 167.71, 180.49, 176.56, 165.79, 163.04,
174.24, 171.99, 172.79, 163.93, 170.52, 173.68, 171.79, 164.07]
X = pd.Series(data)
cls = [160, 165, 170, 175, 180, 185]
freq = X.value_counts(bins=cls).sort_index() # 度数
prop = X.value_counts(bins=cls, normalize=True).sort_index() # 相対度数
cprop = X.value_counts(bins=cls, normalize=True).cumsum().sort_index() # 累積相対度数
freq_tab = pd.DataFrame({
"階級値": freq.index.mid, # 階級区間の中央
"度数": freq,
"相対度数": prop,
"累積相対度数": cprop
}, index=freq.index)
freq_tab
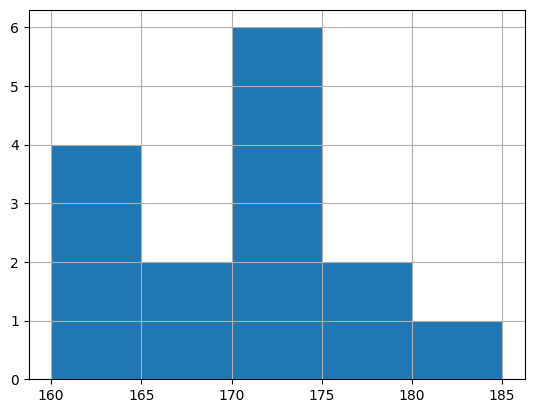
| 階級値 | 度数 | 相対度数 | 累積相対度数 | |
|---|---|---|---|---|
| (159.999, 165.0] | 162.4995 | 4 | 0.266667 | 0.666667 |
| (165.0, 170.0] | 167.5000 | 2 | 0.133333 | 0.800000 |
| (170.0, 175.0] | 172.5000 | 6 | 0.400000 | 0.400000 |
| (175.0, 180.0] | 177.5000 | 2 | 0.133333 | 0.933333 |
| (180.0, 185.0] | 182.5000 | 1 | 0.066667 | 1.000000 |
X.hist(bins = [160, 165, 170, 175, 180, 185])
<Axes: >
度数分布表から平均・分散を求める.#
import numpy as np
import pandas as pd
data = np.array([4, 1, 7, 6, 5, 6, 5, 4, 4, 3, 4, 4, 5, 5, 5, 5, 6, 6, 6, 5, 4, 5, 10])
ds = pd.Series(data)
Xe = np.arange(0, 11) # = [0, 1, ..., 10] # 期待しているユニークなデータセット
f = ds.value_counts().reindex(Xe, fill_value=0).to_numpy() # .to\_numpy()でnumpyの配列に変換
p = ds.value_counts(normalize=True).reindex(Xe, fill_value=0).to_numpy()
print("Xe = ", Xe)
print("f = ", f)
print("p = ", p)
Xe = [ 0 1 2 3 4 5 6 7 8 9 10]
f = [0 1 0 1 6 8 5 1 0 0 1]
p = [0. 0.04347826 0. 0.04347826 0.26086957 0.34782609
0.2173913 0.04347826 0. 0. 0.04347826]
x = np.array([1, 2, 3])
y = np.array([2, 0, 10])
x * y
array([ 2, 0, 30])
Xe * f
array([ 0, 1, 0, 3, 24, 40, 30, 7, 0, 0, 10])
n = X.count()
mean1 = np.sum( Xe * f ) / n
mean2 = np.sum( Xe * p )
print(mean1, mean2)
5.0 5.0
D2 = ( Xe - X.mean() ) ** 2
D2
array([25., 16., 9., 4., 1., 0., 1., 4., 9., 16., 25.])
var1 = np.sum( D2 * f) / n
var2 = np.sum( D2 * p )
print(var1, var2)
2.608695652173913 2.608695652173913
Xe2 = Xe * Xe
Xe2
array([ 0, 1, 4, 9, 16, 25, 36, 49, 64, 81, 100])
Xe2_mean = np.sum( Xe2 * p )
Xe_mean = np.sum( Xe * p)
var = Xe2_mean - Xe_mean ** 2
print(var)
2.608695652173914
np.abs(var1 - var)
np.float64(8.881784197001252e-16)
箱ひげ図#
!pip install japanize-matplotlib
import japanize_matplotlib
Requirement already satisfied: japanize-matplotlib in /usr/local/lib/python3.11/dist-packages (1.1.3)
Requirement already satisfied: matplotlib in /usr/local/lib/python3.11/dist-packages (from japanize-matplotlib) (3.10.0)
Requirement already satisfied: contourpy>=1.0.1 in /usr/local/lib/python3.11/dist-packages (from matplotlib->japanize-matplotlib) (1.3.2)
Requirement already satisfied: cycler>=0.10 in /usr/local/lib/python3.11/dist-packages (from matplotlib->japanize-matplotlib) (0.12.1)
Requirement already satisfied: fonttools>=4.22.0 in /usr/local/lib/python3.11/dist-packages (from matplotlib->japanize-matplotlib) (4.58.4)
Requirement already satisfied: kiwisolver>=1.3.1 in /usr/local/lib/python3.11/dist-packages (from matplotlib->japanize-matplotlib) (1.4.8)
Requirement already satisfied: numpy>=1.23 in /usr/local/lib/python3.11/dist-packages (from matplotlib->japanize-matplotlib) (2.0.2)
Requirement already satisfied: packaging>=20.0 in /usr/local/lib/python3.11/dist-packages (from matplotlib->japanize-matplotlib) (24.2)
Requirement already satisfied: pillow>=8 in /usr/local/lib/python3.11/dist-packages (from matplotlib->japanize-matplotlib) (11.2.1)
Requirement already satisfied: pyparsing>=2.3.1 in /usr/local/lib/python3.11/dist-packages (from matplotlib->japanize-matplotlib) (3.2.3)
Requirement already satisfied: python-dateutil>=2.7 in /usr/local/lib/python3.11/dist-packages (from matplotlib->japanize-matplotlib) (2.9.0.post0)
Requirement already satisfied: six>=1.5 in /usr/local/lib/python3.11/dist-packages (from python-dateutil>=2.7->matplotlib->japanize-matplotlib) (1.17.0)
import numpy as np
import pandas as pd
X = np.array([4, 1, 7, 6, 5, 6, 5, 4, 4, 3, 4, 4, 5, 5, 5, 5, 6, 6, 6, 5, 4, 5, 10])
Y = np.array([9, 6, 1, 6, 5, 10, 3, 2, 7, 2, 5, 8, 7, 6, 1, 8, 4, 5, 4, 0, 9, 3, 4])
df = pd.DataFrame({
'試験X': X,
'試験Y': Y,
})
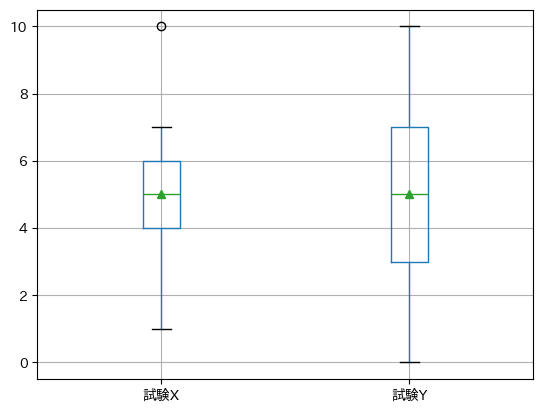
df.boxplot(showmeans=True) # showmeans=Trueで平均値$\bar{x}$が表示される.
<Axes: >
相関#
import numpy as np
np.random.seed(8787)
# 平均, 分散, 標準偏差
mean_X, mean_Y = 170, 68
var_X, var_Y = 5, 10
std_X, std_Y = np.sqrt(var_X), np.sqrt(var_Y)
rho = 0.7 # 相関係数
# 共分散行列
cov = [
[var_X, rho * std_X * std_Y],
[rho * std_X * std_Y, var_Y ]
]
L = np.linalg.cholesky(cov)
# ランダムな非相関標準正規データを生成
num_samples = 1000
uncorr = np.random.standard_normal( (2, num_samples) )
# 相関のあるデータに変換
mean = np.array([mean_X, mean_Y]).reshape(2, 1)
data = np.dot(L, uncorr) + mean
import pandas as pd
df = pd.DataFrame({
"身長[cm]": data[0, :],
"体重[kg]": data[1, :]
})
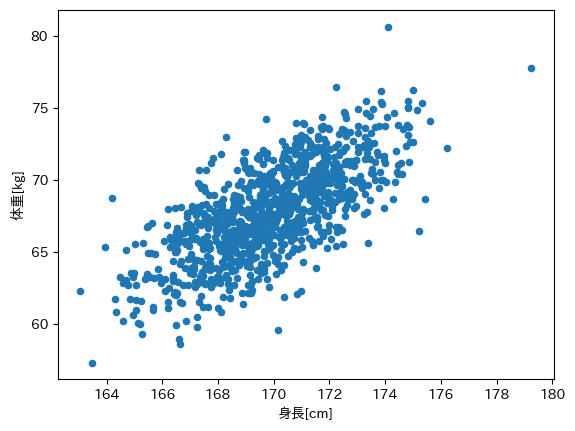
print(df.describe())
身長[cm] 体重[kg]
count 1000.000000 1000.000000
mean 169.841922 67.796619
std 2.267517 3.246836
min 163.060816 57.335005
25% 168.303610 65.626473
50% 169.838044 67.734873
75% 171.444423 70.033005
max 179.247738 80.651467
!pip install japanize-matplotlib
import japanize_matplotlib
Requirement already satisfied: japanize-matplotlib in /usr/local/lib/python3.11/dist-packages (1.1.3)
Requirement already satisfied: matplotlib in /usr/local/lib/python3.11/dist-packages (from japanize-matplotlib) (3.10.0)
Requirement already satisfied: contourpy>=1.0.1 in /usr/local/lib/python3.11/dist-packages (from matplotlib->japanize-matplotlib) (1.3.2)
Requirement already satisfied: cycler>=0.10 in /usr/local/lib/python3.11/dist-packages (from matplotlib->japanize-matplotlib) (0.12.1)
Requirement already satisfied: fonttools>=4.22.0 in /usr/local/lib/python3.11/dist-packages (from matplotlib->japanize-matplotlib) (4.58.4)
Requirement already satisfied: kiwisolver>=1.3.1 in /usr/local/lib/python3.11/dist-packages (from matplotlib->japanize-matplotlib) (1.4.8)
Requirement already satisfied: numpy>=1.23 in /usr/local/lib/python3.11/dist-packages (from matplotlib->japanize-matplotlib) (2.0.2)
Requirement already satisfied: packaging>=20.0 in /usr/local/lib/python3.11/dist-packages (from matplotlib->japanize-matplotlib) (24.2)
Requirement already satisfied: pillow>=8 in /usr/local/lib/python3.11/dist-packages (from matplotlib->japanize-matplotlib) (11.2.1)
Requirement already satisfied: pyparsing>=2.3.1 in /usr/local/lib/python3.11/dist-packages (from matplotlib->japanize-matplotlib) (3.2.3)
Requirement already satisfied: python-dateutil>=2.7 in /usr/local/lib/python3.11/dist-packages (from matplotlib->japanize-matplotlib) (2.9.0.post0)
Requirement already satisfied: six>=1.5 in /usr/local/lib/python3.11/dist-packages (from python-dateutil>=2.7->matplotlib->japanize-matplotlib) (1.17.0)
df.plot(kind='scatter', x='身長[cm]', y='体重[kg]')
<Axes: xlabel='身長[cm]', ylabel='体重[kg]'>
import numpy as np
import matplotlib.pyplot as plt
np.random.seed(87)
ns = 1000 # number of samples
def gen_corr_data(ns, rho):
# 平均, 分散, 標準偏差
mean_X, mean_Y = 170, 68
var_X, var_Y = 5, 10
std_X, std_Y = np.sqrt(var_X), np.sqrt(var_Y)
rho = rho # 相関係数
# 共分散行列
cov = [
[var_X, rho * std_X * std_Y],
[rho * std_X * std_Y, var_Y ]
]
L = np.linalg.cholesky(cov)
# ランダムな非相関標準正規データを生成
uncorr = np.random.standard_normal( (2, num_samples) )
# 相関のあるデータに変換
mean = np.array([mean_X, mean_Y]).reshape(2, 1)
data = np.dot(L, uncorr) + mean
return data
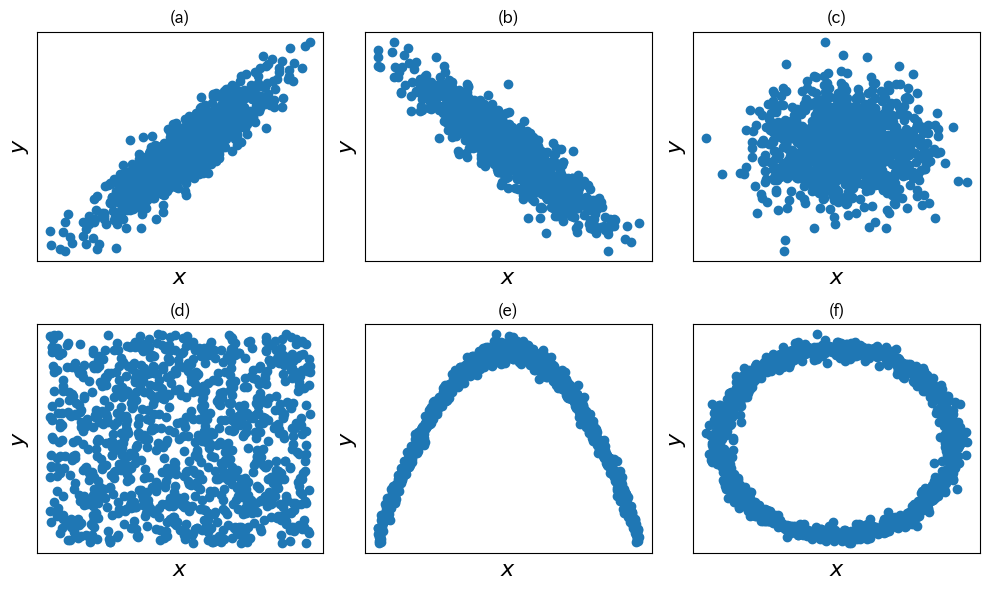
fig, ax = plt.subplots(2, 3, figsize=(10,6))
X1, Y1 = gen_corr_data(ns, 0.9)
X2, Y2 = gen_corr_data(ns, -0.9)
X3, Y3 = gen_corr_data(ns, 0)
X4, Y4 = np.random.random(ns), np.random.random(ns)
x = np.linspace(-10, 10, ns)
eps = np.random.normal(0, 3, ns) # noise
y5 = - x ** 2 + eps
eps = np.random.normal(0, 0.05, ns) # noise
z = (1 + eps)*np.exp( 1.j * 2*np.pi * x)
ax[0,0].scatter(X1, Y1)
ax[0,1].scatter(X2, Y2)
ax[0,2].scatter(X3, Y3)
ax[1,0].scatter(X4, Y4)
ax[1,1].scatter(x, y5)
ax[1,2].scatter(z.real, z.imag)
#ax[3].scatter(z.real, z.imag)
tit=["(a)", "(b)" ,"(c)", "(d)", "(e)", "(f)"]
for i, a in enumerate(ax.flat):
a.set_xlabel(r"$x$", fontsize=16)
a.set_ylabel(r"$y$", fontsize=16)
a.set_title(tit[i])
a.set_xticks([])
a.set_yticks([])
plt.tight_layout()
for i,j in